What is the Common Workflow Language?
The Common Workflow Language (CWL) is a widely supported, open source, vendor neutral specification for workflow descriptions. Because there are a wide variety of executors that support CWL, the same CWL document will run on a laptop, on HPC clusters, on raw cloud infrastructure and on a variety of cloud platforms.
See the current list of implementations that support CWL here.
Looking for how to implement something in CWL? This list of CWL Patterns might help.
What does Rabix provide?
Rabix Benten: Code intelligence for CWL editing
Benten is a language server for Common Workflow Language documents. It works with any editor/IDE that has language server support, such as VS Code, VI, emacs etc.
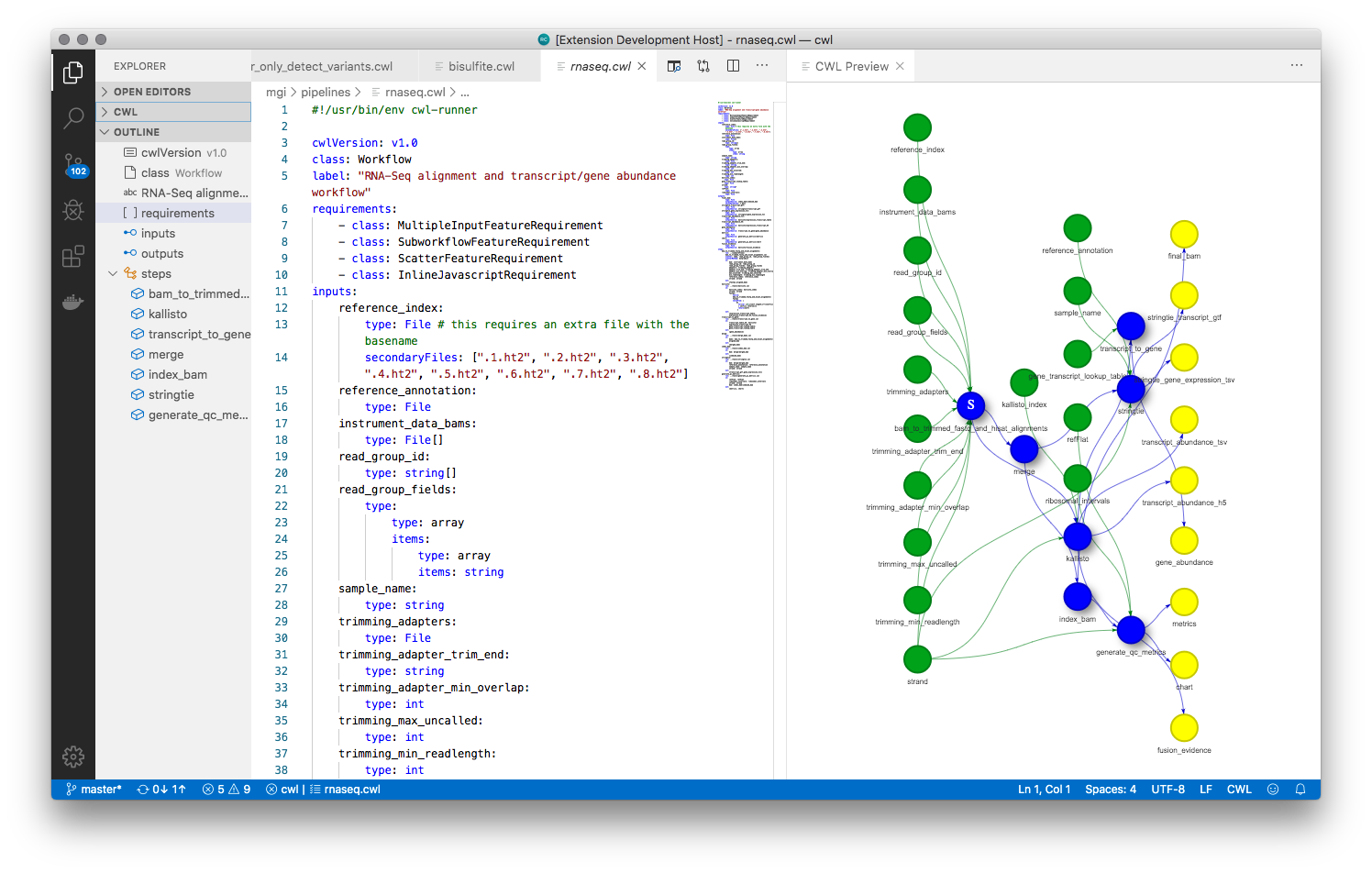
sbpack: Command line interface for pushing and pulling local CWL to/from a Seven Bridges Platform
sbpack is a command line tool, written in Python, to upload (sbpack) and download (sbpull) CWL apps to/from any Seven Bridges powered platform. This enables local text-based CWL development, use of version control for workflows, and the ability to send workflows to and from the platform with ease.
CWL-SVG: interactive SVG visualization of CWL workflows
CWL-SVG is a Typescript library for visualization of Common Workflow Language workflows. It can be easily integrated into a website or typescript application to show interactive views of a CWL workflow.
CWL-SVG is used in variety of projects including Data Science and Bioinformatics Research Group's Omics Dashboard, Arvados workbench, Vue CWL, the Scientific Data Analysis Platform, and, of course, in Rabix Composer which is used on the Seven Bridges platform.
CWL-TS: Typescript data model for Common Workflow Language
CWL-ts is a data model library for CWL made for use in TypeScript and JavaScript (primarily client-side) applications. It supports JSON schemas and mappings for draft-3, draft-4, and v1.0. Draft-2 support is SevenBridges flavored.
Rabix Composer: Visual programming in CWL [Legacy]
CWL documents are text files describing computational steps. CWL written in YAML format is compact and easily read by users. While some users are comfortable creating CWL workflows "by hand" using a text editor, others may prefer Rabix Composer: a powerful, open source, graphical editor allowing visual programming in CWL. Rabix Composer enables a user to quickly create complex CWL workflows without having to remember the details of CWL syntax.
Support Notice
Rabix Composer will remain available as a local sbg:draft2 and CWL 1.0 tool and workflow editor. However, please note that Seven Bridges is not actively maintaining or extending Rabix Composer at the moment (Q2 2021). The repository will remain open for new issues and pull requests, but please have in mind that those may not be processed in a timely manner.
If you need an up-to-date CWL editor, you can create an account either on the CGC, Biodata Catalyst powered by Seven Bridges, or on CAVATICA and use the Web Composer available there. Web Composer is also available on the Seven Bridges Commercial offerings.

Rabix Executor: Runner for CWL pipelines [Legacy]
Rabix Executor is a workflow runner that can execute CWL sbg:draft2 and CWL 1.0 pipelines. It has support for the TES API and can be used in a limited manner to run pipelines on cloud infrastructure. [PubMed, bioRxiv]
When the Rabix project was in it's infancy and the CWL specification was just being developed (ca. 2014-2016) Rabix Executor was the only executor capable of running CWL. It proved invaluable as a test bed for ideas for the specification that evolved into CWL and as a living implementation of the specification.
There are now many modern CWL executors for every conceivable platform. Rabix Executor is now in maintenance mode. Rabix Executor remains a stable local test runner for those people wishing to run sbg:draft2 pipelines locally, but no new features will be added to it.
With love from Seven Bridges
Seven Bridges is passionate about open standards in science and about providing high quality open source developer tooling around those standards. Rabix (a portmanteau of "Reproducible Analyses for Bioinformatics") was founded by SevenBridges in 2015 to support this mission. The Rabix team plays a key role in developing CWL and tooling around CWL.